Ancient proteins may offer clues on how to engineer proteins that can withstand the high temperatures required in industrial applications, according to new research published in the Proceedings of the National Academy of Sciences. Researchers used experiments to examine critical differences between 15 proteins from the thioredoxin family, including seven extinct protein sequences that date back more than 4 billion years and were resurrected using ancestral sequence reconstructions.
Ancient proteins may offer clues on how to engineer proteins that can withstand the high temperatures required in industrial applications, according to new research published in the Proceedings of the National Academy of Sciences. Researchers used experiments to examine critical differences between 15 proteins from the thioredoxin family, including seven extinct protein sequences that date back more than 4 billion years and were resurrected using ancestral sequence reconstructions.
“In industrial applications, proteins could do some of the work of chemicals with less energy and less toxicity, but they have to be able to withstand the higher temperatures of industrial processes,” said George Makhatadze, a Constellation Professor of Biocomputation and Bioinformatics and member of the Center for Biotechnology and Interdisciplinary Studies. “This research suggests that ancestral reconstruction methods can be used to design proteins that unfold more slowly, offering greater utility, at higher temperatures.”
The experiments showed that the difference in stability between proteins from the same family is caused primarily by differences in the energy required to unfold the proteins, a finding that upholds a long-held theory known as the “principle of minimal frustration,” proposed by physicists about 30 years ago.
Proteins are molecular chains in which each link is one of 20 amino acids. Once the chain is assembled, various forces along its length interact, causing the string to twist and turn, and ultimately fold into a three-dimensional shape. The protein can only perform its function when properly folded. Most proteins remain folded within a set range of temperature, pH, or pressure conditions, breaking down when subjected to conditions outside of these tolerances.
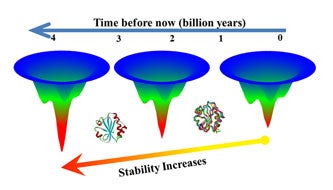
Not all the rules that govern protein folding are known, and Makhatadze wanted to understand how proteins within the same family, with similar structure, are able to perform the same life-sustaining function in vastly different thermal environments. Thioredoxins, for example, are present in all organisms, from those that live near boiling hydrothermal vents to frigid Arctic waters.
One known rule helps to explain why there are an astronomical number of sequences possible for proteins with hundreds and even thousands of links, but only a small subset of those possible sequences are found in nature. This rule, the principle of minimal frustration, posits that nature chooses only those sequences that fold most efficiently, reducing undesired interactions between amino acids and producing faster, less frustrated, folding patterns.
To be consistent with the principle of minimal frustration, Makhatadze said, proteins with similar structure but different thermodynamic stability should fold along the same efficient pattern, but those with greater ability to tolerate heat should take longer to unfold. Experiments, undertaken first with eight modern-day thioredoxins, proved the hypothesis correct.
Previous research supports the finding that older proteins are more stable, but this new research shows these ancient proteins are more stable because they unfold more slowly.—Makhatadze
“We tested the folding rates among thioredoxins, a family of proteins that perform the same function but—because they function in different thermal conditions—must be different in terms of thermodynamics,” said Makhatadze. “And what we observed is that for these proteins, they indeed fold with the same rate, but they unfold with a different rate—which is what we predicted.”
In the next step, researchers tested extinct versions of thioredoxins that had been obtained using a technique called ancestral sequence reconstruction. The extinct versions unfolded more slowly than modern versions, in some case folding 3,000 times more slowly than a modern counterpart.
“Here we have two versions of a protein from the same family and one unfolded in seven seconds, the other in six hours,” Makhatadze said. “That means you can get useful function from the protein for six hours versus seven seconds. This shows that ancestral sequence reconstruction may be a route to proteins that are more stable at high temperatures.”
The finding is supported by previously published research and geochemical data which indicates that Earth and its oceans were hotter in the ancient past.
“Previous research supports the finding that older proteins are more stable, but this new research shows these ancient proteins are more stable because they unfold more slowly,” Makhatadze said.
“Evidence for the principle of minimal frustration in the evolution of protein folding landscapes” can be found using the digital object identifier doi: 10.1073/pnas.1613892114. The research was supported by the National Science Foundation. Makhatadze was joined in the research by Franco Tzul, a postdoctoral research associate, and graduate student Daniel Vasilchuk.